The microbial world influences almost every aspect of our lives. Microbes live in diverse communities made up of multiple species that work together and impact each other, whether they are in the soils where our food is grown, the lungs of a person with an infection, or at the bottom of the ocean. The geography of how a microbial community is laid out affects how those microbes live and function together, just as it does in our own neighborhoods.
Caltech researchers have discovered that changes in local oxygen concentration have a significant impact on whether microbial neighbors live or die in the presence of nitric oxide, a common microbial by-product (NO). The findings imply that large-scale global models, such as those of the nitrogen cycle, should strive to incorporate the fact that chemical microscale environments influence microbial behavior.
On October 27, a paper describing the research will be published in the journal Current Biology. The research was led by graduate student Steven Wilbert and conducted in Dianne Newman's laboratory, Gordon M. Binder/Amgen Professor of Biology and Geobiology and executive officer for biology and biological engineering.
Nitric oxide is created as a byproduct of the multistep process of converting nitrate (NO3-) to nitrogen gas (N2). This entire process, known as denitrification, is an essential component of biological processes all over the world. According to recent research, different steps in this pathway may be performed by members of various microbial communities.
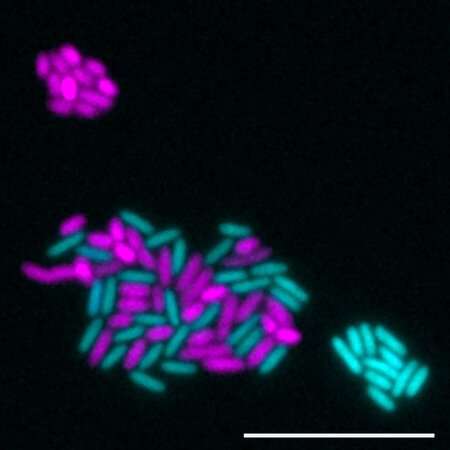
Wilbert used Pseudomonas aeruginosa, a bacterium extensively studied in the Newman laboratory, as a model organism to investigate how a microbe's local environment affects its ability to carry out the denitrification process. Wilbert created a strain that only executed the first half of the denitrification pathway and another strain that only executed the second half of the pathway using genetic engineering techniques.
Wilbert then investigated how those two engineered bacterial strains interact in different oxygen environments. The idea was that the "first half" strain produces NO as a byproduct, and the team wanted to know how the "second half" strain would deal with the NO under different local oxygen concentrations, and how this would affect the entire community.
In the absence of oxygen, the second genetically engineered strain of Pseudomonas was able to take NO produced by the first and chemically alter, or reduce, that chemical as part of the normal denitrification process, according to the findings. Furthermore, the bacteria were able to grow on NO as a substrate. However, in a higher oxygen environment, NO became toxic, killing Pseudomonas strains that couldn't reduce the molecule.
"Oxygen dramatically tunes these microbial interactions: they can either live or die as a result," says Newman. "This, in turn, has an impact on the entire denitrification process. Models attempting to account for how microorganisms contribute to the nitrogen cycle must therefore account for the microscopic spatial environment. This is a critical variable."
While the study revealed specifics about how oxygen mediates cellular interactions with NO, it also points to more general principles about a wide range of microbial by-products. NO is an example of a RAM (redox-active metabolite). This research opens up a new avenue for investigating how RAMs' effects on microbes are influenced by their local microenvironment, which can vary greatly in space and time.
"Microbial metabolism is a race to pick up and drop electrons," Wilbert explains. "Everything in life revolves around the transfer of energy. RAMs, with their ability to donate and receive electrons, are valuable currency among microbial neighbors. While this can help with energy transfer, RAMs are sensitive to changes in local oxygen concentrations that vary over time and space. Because of our NO results, we're convinced that oxygen is the key to gaining a better understanding of what's going on in unseen soils, oceans, and anywhere else microbial interactions may occur. We can make better predictions of how microbial communities survive in the lungs or agricultural systems if we understand how oxygen changes at the microscale level."
"The contrasting roles of nitric oxide drive microbial community organization as a function of oxygen presence," the paper says.
Journal information: Current Biology